|
xx |
xx |
|
 |
 |
|
INFECTIOUS
DISEASE |
BACTERIOLOGY |
IMMUNOLOGY |
MYCOLOGY |
PARASITOLOGY |
VIROLOGY |
|
DNA tumor viruses have been moved to a separate page
Follow Previous below |
VIROLOGY - CHAPTER SIX
PART TWO
RNA TUMOR VIRUSES
Retroviruses
Dr Richard Hunt
Professor
Department of Pathology, Microbiology and Immunology
University of South Carolina School of Medicine
|
|
|
|
EN
FRANCAIS |
|
En
Español |
|
NË SHQIPTARE |
|
|
|
Let us know what you think
FEEDBACK |
|
SEARCH |
|

 |
|
|
|
|
|
Figure 10
 Retrovirus
replication
Retrovirus
replication
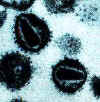 Human
immunodeficiency virus
Copyright Department of Microbiology, University of
Otaga, New
Zealand.
Human
immunodeficiency virus
Copyright Department of Microbiology, University of
Otaga, New
Zealand.
 Figure 11
Figure 11
Structure of a retrovirus: (The virus shown is
human immunodeficiency virus-1) From the Harvard AIDS Institute Library of Images, courtesy of Critical
Path AIDS Project, Philadelphia.
|
RNA tumor viruses are different from DNA tumor viruses in that their genome is RNA
but they are similar to many DNA tumor viruses in that the genome is integrated into
the host
genome.
Since RNA makes up the genome of the mature virus particle, it must be
copied to DNA prior to integration into the host cell chromosome. This
life style (figure 10) goes against the central dogma of molecular biology in which that DNA is
copied into RNA.
RNA tumor viruses or
oncornaviruses are members of the retrovirus family.
Retrovirus structure
The outer envelope comes from the host cell plasma membrane
(figure 11).
Coat proteins (surface antigens) are encoded by
env
(envelope) gene and are glycosylated. One primary gene product is made but this is cleaved so that
there are more than one surface glycoprotein in the mature virus (cleavage is by host
enzyme in the Golgi apparatus). The primary protein (before cleavage) is made on
ribosomes attached to the endoplasmic reticulum and is a transmembrane (type 1)
protein.
Inside the membrane is an icosahedral capsid
containing proteins encoded by the gag
gene (Group- specific AntiGen). Gag- encoded proteins also
coat the genomic RNA. Again there is one primary gene product. This is cleaved
by a virally-encoded protease (from the pol gene)
There are two molecules of genomic RNA per virus
particle with a 5' cap and a 3' poly A sequence. Thus, the virus is diploid. The
RNA is plus sense (same sense as mRNA).
About 10 copies of reverse transcriptase are
present within the mature virus, these are encoded by the pol
gene.
Pol
gene codes for several functions (again, as with
gag and env, a polyprotein is made that is then cut up)
|
 Figure 12
Figure 12
Structure of RSV protease bound to a peptide analog of the HIV cleavage
site
Requires Chime plug-in.
|
The pol gene products are:
a) Reverse transcriptase (a polymerase that copies RNA to DNA)
b) Integrase (integrates the viral genome into the host genome)
c) RNase H (cleaves the RNA as the DNA is transcribed so that reverse
transcriptase can make the second complementary strand of DNA)
d) Protease (cleaves the polyproteins translated from mRNAs from the
gag
gene and the pol gene itself). This virally encoded protease is
the target of a new generation of anti-viral drugs (figure 12).
|
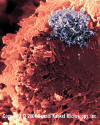 Figure 13 Figure 13
Human T lymphocyte with HTLV-1 infection
(RNA virus, Retroviridae Family). The virus is in a large clump in the
corner
©
Dennis Kunkel Microscopy, Inc.
Used with permission
|
GROUPS OF RETROVIRUSES
Oncovirinae
These are the tumor viruses and those with similar morphology. The
first member of this group to be
discovered was Rous Sarcoma Virus (RSV) - which causes a slow neoplasm in chickens.
Viruses in this group that cause tumors in humans are:
HTLV-1 (human T-cell lymphotropic virus-1, figure
13)
which causes Adult T-cell Leukemia (Sezary T-cell
Leukemia). This disease is found in some Japanese islands, the Caribbean, Latin America and Africa. HTLV-1
is sexually transmitted
HTLV-2 (human T-cell lymphotropic virus-2)
which causes Hairy Cell Leukemia. The virus is endemic to very specific regions
of the Americas, particularly in native American populations.
Lentivirinae
These have a long latent period of infection
before disease occurs; they were mainly associated
with diseases of ungulates (e.g. visna virus) but HIV (formerly HTLV-III) which
causes AIDS belongs to this group. It is much more closely related to some Lentivirinae
than it is to HTLV-I and HTLV-II which are Oncovirinae.
Spumavirinae
There is no evidence of pathological effects of these viruses. They
establish persistent infections in many animal species. They have been isolated
from primates (including humans), cattle, cats, hamsters, and sea lions.
Cells infected by spumaviruses have a foamy appearance (because of numerous
vacuoles) and often form
syncytia of
giant multinucleate cells. Chimpanzee (simian) foamy virus is the type species. Human
foamy virus is a variant of simian foamy virus and is usually acquired from a
monkey bite.
|
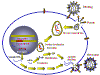 Figure 14
Figure 14
Stages in the productive infection of a cell by a retrovirus
|
INFECTION AND TRANSFORMATION OF A CELL BY A RETROVIRUS
The following stages occur in the infection process
(figure 14):
- Binding to a specific cell surface receptor
- Uptake by endocytosis or by direct fusion to the plasma membrane.
The virus may require entry into a low pH endosome before fusion can occur, although some
(e.g. HIV) can
fuse directly with the plasma membrane
- RNA (plus sense) is copied by reverse transcriptase to minus sense DNA. Here,
the polymerase is
acting as an RNA-dependent DNA polymerase. Since reverse transcriptase is a DNA
polymerase, it needs a primer. This is a tRNA that is incorporated into the
virus particle from the previous host cell.
- RNA is displaced and degraded by a virus-encoded RNase H activity.
Reverse transcriptase now acts as a DNA-dependent DNA
polymerase and copies the new DNA into a double strand DNA. This DNA form of the
virus is known as a the provirus.
- Double strand DNA is circularized
and integrated into host cell DNA (see
below) using a virally encoded integrase enzyme. This DNA is copied every time cellular DNA is copied. Thus, at this stage the
provirus is just like a normal cellular gene.
- Full length, genomic RNA (plus sense) is copied from
the integrated DNA by host RNA
polymerase II which normally copies a gene to mRNA. The genomic RNA is capped and poly adenylated,
just as an mRNA would be.
Since the full length genomic RNA is the same sense as message, it also
acts as the mRNA for GAG and POL polyproteins.
The genomic RNA is spliced by host nuclear enzymes to give mRNA for other proteins such
as ENV. The RNA of some more complex retroviruses such as HTLV-1 and HIV undergoes
multiple splicing (see chapter 7, HIV).
Note that mRNA comes from splicing genomic RNA or is the genomic
RNA. As a result, both mRNA and genomic RNA must be the same sense
- since mRNA must
be plus sense, the genomic RNA of all retroviruses must also be plus sense.
An advantage of this mode of replication is that it allows growth in terminally
differentiated cells since the only host cell polymerase usurped by the virus is RNA
polymerase II which is present in all cells.
|
| |
MECHANISM OF VIRAL GENOME REPLICATION
If host RNA polymerase II is used to copy the DNA back to RNA,
there are major problems with having a DNA provirus form but an RNA genome in
the mature virus particle
These problems include:
- RNA polymerase II does not copy the upstream and down
stream control sequences of genes. It only copies the information necessary to
make a protein
- The lack of proof reading by RNA polymerase II
Failure of RNA polymerase II to copy the entire gene
The problem is that, when transcribing genes, RNA polymerase II needs control and
recognition sites upstream from the transcription initiation
site. The upstream site at which the polymerase molecule binds is called the PROMOTOR.
Promotors are not themselves copied into mRNA since they have no function in the
translation of protein. After binding to the promotor, the polymerase begins
transcription at a downstream site, the RNA initiation site. The polymerase
continues to transcribe DNA into RNA until it reaches a termination/polyadenylation
signal, part of which is not copied since,agaun, it also has no function
in the making of the protein. Furthermore, both up and downstream from the transcribed
region are control sequences that modulate the transcription of the gene. These are called ENHANCERS.
These are essential parts of any gene and must be present for
RNA polymerase II to work but they are not copied to RNA. This is because RNA
polymerase II in the host cell has the function of making messenger RNA which is
dispensed with after translation. To make a protein, the actual mRNA molecule does not
need the control sequences of the original gene. Thus, the use of host RNA polymerase
II means that the control sequences in the original genome should not get into the RNA
genome of progeny virions.
This means that either the DNA copy of the viral RNA genome virus must integrate
into host DNA downstream from a host promotor and upstream from host termination sites
(a tall order indeed!) or it must find a way of providing its own control
sequences (which, as we said, are not copied into progeny genome). It does the latter in a
most complex manner.
|
Figure 15
A
 The structure of the RNA genome of the mature retrovirus
The structure of the RNA genome of the mature retrovirus
B
 The genome structure of the DNA proviral form of a retrovirus
The genome structure of the DNA proviral form of a retrovirus
|
How can a retrovirus provide its own control promotors and enhancers if they are not
transcribed when the DNA provirus is copied to the genomic RNA form?
Here is a brief (and very incomplete) summary of how a retrovirus does it:
- The viral RNA is composed of three regions. At each end are repeats (called, not
surprisingly, terminal repeats). The repeat sequences (R) (shown in green in
figure 15) do not
code for proteins. In between the two repeats, there is a unique (not repeated) region
that contains the viral genes that code for the proteins (GAG, POL and ENV) plus
other unique sequences at either end that do not code for protein. Near the 5' end
of the RNA genome is the U5 region and near the 3' end is the U3 region. PBS (in
figure 15) is the primer binding site. The tRNA binds here when
reverse transcriptase
starts copying the RNA. PPT is a polypurine tract.
- In the integrated form (when transcribed into DNA and inserted into the host
cell chromosome), the provirus is more complicated. We find that part of the 3'
unique region (called U3) of the RNA genome has been copied and transposed to the opposite
end of the genome. Conversely, part of the 5' end of the unique region (called U5) has been copied and
transposed to the other end. This gives the integrated DNA the structure shown
in figure 15B. For convenience, only one strand of the DNA is shown.
|
|
MOVIE
LTR formation
Requires
Flash
|
Now, of course, there are larger
terminal repeats since the U3 and U5 regions are also repeated. The U3-R-U5 regions
are known as long terminal repeats or LTRs. The U3 region contains all of
the promotor information that is necessary to start RNA transcription at the beginning
of the R (repeat) region while the U5 region contains all of the information necessary
to terminate after the other R repeat. In addition, the LTRs contain information that
enhances the degree of transcription of the three retroviral genes (enhancer regions).
These enhancers can be up or downstream from the protein-encoding part of
the genes.
|
| |
 Figure 16
Figure 16
The transcription of a retrorviral DNA with LTRs by RNA
polymerase II results in the loss of the LTRs
Animated version here |
Host RNA polymerase II copies the provirus DNA to genomic RNA which can be also spliced
to mRNAs. Since the polymerase starts after the promotor (in U3), at the
transcription initiation site, it begins exactly at the beginning of the R
region (figure
16).
Thus we get a faithful (almost - see below) copy of the RNA that entered the cell. The
termination sequences and poly A signal are in U5 which is also not copied.
Because of this mechanism, there can be only one promotor site (from U3)
for all three viral genes so they must be all transcribed together. Splicing
enzymes, from the host cell nuclear splicing machinery, cut the primary
transcript to form the individual mRNAs where
necessary. (See chapter 7,
HIV in which this has been well
elucidated). Unlike the
situation with DNA tumor viruses, there is no distinction between early/late functions.
You may ask why, if U5 contains the termination and polyadenylation sites, does
the transcript not just terminate at the end of the first R region of the LTR
(figure 15b) and never
get into the structural genes. The termination site in the first U5 is suppressed, often
by complex secondary structure mechanism. In some retroviruses there is a sequence in the
gag gene that provides the context to suppress the termination activity of the first U5.
Clearly
the second U5 does not have a gag gene following it.
This strategy of virus replication in which viral RNA is first copied to DNA (by
reverse transcriptase) which then gives rise to mRNA and protein poses another problem for the
virus. The initial step (RNA to DNA) is carried out by a viral
enzyme which is not normally in the cell. Yet this transcription step must occur before
any mRNA transcription or protein translation can occur. The problem is solved by the
virus carrying about 10 copies of the reverse transcriptase protein into the cell with it.
These were packaged when the virus was assembled in the previous host cell. In
theory, the viral genomic RNA coming into the cell could act as an mRNA but it
is too coated with protein to do so. Thus, new mRNA must be made which requires
the synthesis of reverse transcriptase.
|
| |
|
 Figure 17
Figure 17
Typical retrovirus structure and the structure of a retrovirus with an
oncogene (Rous Sarcoma Virus)
|
ONCOGENES IN RETROVIRUSES
The structure shown in figure 15A and the upper part of figure 17 is that of a typical retrovirus with three structural
genes (gag, pol and env) but none of these is oncogenic. If the virus is to transform a cell,
it must have sequences that alter
cellular DNA synthesis and provide the other functions that are typical of a transformed cell.
These are in addition to the gag/pol/env genome.
Thus we also find an ONCOGENE (onc) in the viral genome of many retroviruses
that transform cells to neoplasia (figure 17). It should be emphasized that the
oncogene in RNA tumor viruses is not necessary for viral replication. It is an
additional gene that gives the virus its capacity to transform the host cell.
Definition of virally-induced transformation: The changes in the biologic function
and antigenic specificity of a cell that result from integration of viral genetic
sequences into the cellular genome and that confer on the infected cell certain properties
of neoplasia. Note, however, that transformation can be induced by factors other than
viruses e.g. carcinogens.
What are the oncogenic genes in
retroviruses?
In retroviruses, these were first discovered as an extra gene in Rous sarcoma
virus (RSV) (figure 17). This gene was called src (for sarcoma).
src is not needed for viral replication. It is an extra gene to those (gag/pol/env)
necessary for the continued reproduction of the virus. RSV has a complete gag/pol/env
genome. Deletions/mutations in src abolish transformation and tumor promotion but
the virus is still capable of other functions. RSV is unusual in that it has managed to
retain its whole genome of gag/pol/env.
|
| |
In sharp contrast to RSV, many retroviruses have
lost part of their genome to accommodate an oncogene (figure 18). This has two consequences:
- The protein encoded by the oncogene is often part of a fusion protein with other virally-encoded amino acids
attached
- Virus is in trouble as it cannot make all of itself. To replicate and bud from the host
cell needs products of another virus, that is a helper virus.
About forty oncogenes have now been identified. Note that they are referred so by a
three letter code (e.g. src, myc) often reflecting the virus from which they were
first isolated. Some viruses can have more than one oncogene (e.g. erbA,
erbB).
Because they are viral oncogenes, we put a letter v in front of the three
letter name. Here are a few of the most studied:
|
Virus |
Oncogene |
|
Rous sarcoma virus |
v-src |
|
Simian sarcoma virus |
v-sis |
|
Avian erythroblastosis virus |
v-erbA or v-erbB |
|
Kirsten murine sarcoma virus |
v-kras |
|
Moloney murine sarcoma virus |
v-mos |
|
MC29 avian myelocytoma virus |
v-myc |
|
| |
CELLS HAVE PROTO-ONCOGENES
Once retroviral oncogenes had been discovered, a surprising observation was made:
Unlike the situation with DNA virus oncogenes which are true viral genes,
there are homologs of all retrovirus oncogenes in cells that are not infected by a
retrovirus. These cellular homologs are often genes involved in growth control
and development/differentiation (as might be expected) and have important
non-transforming functions in the cell; some can cause cancer under certain circumstances
and, presumably, those not shown to cause cancer have the ability to do so under the
correct conditions. The cellular homologs of viral oncogenes are called proto-oncogenes.
To distinguish viral oncogenes from cellular proto-oncogenes, they are often referred to
as v-onc and c-onc respectively. Note that c-oncs are not identical to their
corresponding v-oncs. It appears that the virus has picked up a cellular growth
controlling or differentiation gene and, after the gene was acquired by the virus, it has been subject to
mutation.
Definition of a proto-oncogene: A host gene that is homologous to an
oncogene that is
found in a virus but which can induce transformation only after being altered (such as
mutation or a change of context such as coming under the control of a highly active
promotor). It usually encodes a protein that functions in DNA replication or growth
control at some stage of the normal development of the organism.
Characteristics of cellular proto-oncogenes
- These are typical cellular genes with typical control sequences. As with most
eucaryotic genes, most have introns (while retroviral oncogenes - v-oncs - do not)
- They show normal Mendelian inheritance because
they are normal genes, essential to the functions of the cell.
- As with all genes in the eukaryotic genome, they are always at same place in genome
(cf. what would be expected of endogenous retroviruses that had, over time, become
incorporated into the cellular genome)
- There are no LTR sequences (v-oncs always are in an LTR context)
- Viral oncogenes are most like the c-onc of the animal from which the virus is thought to have
acquired the gene. Thus, v-src of RSV is more like chicken src than human src. Note
that v-onc
was long ago acquired accidentally by the virus from the genome of a previous host cell
- Cellular oncogenes are expressed by the cell at some period in the life of
the cell, often when the
cell is growing, replicating and differentiating normally. They are usually proteins
that are involved
in growth control.
- Cellular oncogenes are highly conserved
If v-onc and c-onc are so alike, why does the v-onc introduced by a
virus cause havoc in the cell? This is due to differences in the genes, mutations that have occurred
in the gene once it was picked up by the virus. Such changes include:
- Amino acid substitutions or deletions which
result in altered translation products
- Many v-onc proteins are fusion proteins
translated from a v-onc that is a hybrid gene of a c-onc and a viral
gene.
- V-oncs are inserted into the host genome along with LTRs which
contain promotors/enhancers. This is likely
to result in over expression of a gene that we know is probably involved in control
of DNA transcription and replication!
|
 Figure 18
Figure 18
Some retroviruses that have an oncogene that
replaces their normal genes
|
Chronically transforming retroviruses
do not have a v-onc
The observation that an acutely transforming virus such as RSV contains an extra
gene, the oncogene, explains their high neoplastic potential but, in contrast, chronically-transforming retroviruses only produce tumors slowly and they carry no gene
equivalent to a v-onc. At best, these viruses have just the three usual viral genes (gag/pol/env). An
example is avian leukosis virus (ALV) (figure 18).
How do chronically transforming viruses induce a tumor if they do not have an oncogene?
A seminal observation was made: Just as any other retrovirus does, ALV can integrate
into the cell genome at many different sites but, in ALV-induced tumors, the virus
is ALWAYS found in a similar position (very important!). This means that the
crucial transforming event must be rare and that the cells that form the tumor are a clone
(cf. the acute transformers which are found all over the place). In all cases of ALV-induced tumors, the viral genome is inserted near a cellular gene called
c-myc.
This is the cellular proto-oncogene that, in an altered form (i.e. as a
v-onc), is
carried by some acutely transforming retroviruses (e.g. avian myelocytoma virus
which causes carcinoma, sarcomas and leukemias). In addition, the level of translation of
c-myc in the ALV-transformed cell is much greater than in uninfected cells. Thus,
inserting the genome of ALV or other chronically transforming retroviruses next to a
c-onc has the same effect as carrying in a v-onc.
|
 Figure 19
Figure 19
Oncogenesis by
promotor insertion
 Figure 20
Figure 20
Oncogenesis by enhancer insertion
|
So, during integration, the virus comes to lie upstream from
c-myc which then comes
under the influence of the strong LTR promotors of the virus which leads to over
expression of c-myc. This is called oncogenesis by promotor insertion
(Figure 19).
But in some tumors the virus is downstream from the
c-myc gene.
However, we saw that LTRs also have enhancers in addition to promotors. We know
that enhancer sequences can be upstream or downstream to have their effect. This is called
oncogenesis by enhancer insertion (Figure 20).
Why is insertion near c-myc important? The protein coded for by this gene is found in
the nucleus of normal cells and is involved in control of DNA synthesis. It can be shown
that over-expression of c-myc leads to rapid DNA replication.
|
|
 Figure 21A
Figure 21A
Many genes can be assigned to sites on specific chromosomes
 Figure 21B
Figure 21B
Many break sites in chromosomes are very close to
a cellular proto oncogene
|
Can cellular ongogenes be involved in non-virally
induced cancer?
Once it had been shown that viruses can either bring an oncogene into the cell
or can take control of a cellular proto-oncogene to give rise to a tumor, the question arose of whether cellular
proto-oncogenes could give rise to tumors in the absence of retroviral infection. The
answer is yes! Other chromosomal rearrangements can bring a c-onc under the control
of the wrong promotor/enhancer (Figure 21). Alternatively, the c-onc might be mutated in a particular
way so that it was over-expressed or it might code for a mutant protein with an altered
function.
Chromosomal mapping allows the precise localization of the site of a gene on a
particular chromosome and many cancers are associated with alterations in chromosomes,
particularly translocations (the breakage of a chromosome so that the two parts associate
with two parts of another chromosome).
Many break sites in tumor cells are very
close
to a known c-onc. This is highly suggestive and unlikely to have occurred by chance!
| Disease |
C-onc |
translocation |
Burkitt's lymphoma * |
myc |
8 to 14 |
Acute myeloblastic leukemia |
mos |
8 to 21 |
Chronic myelogenous leukemia |
abl |
9 to 22 |
Acute promyelocytic leukemia |
fes |
15 to 17 |
Acute lymphocytic leukemia |
myb |
6 deletion |
Ovarian cancer |
myb |
6 to 14 |
* In Burkitt's lymphoma the
c-myc on chromosome 8 is brought to a
site on chromosome 14 close to the gene for immunoglobulin heavy chains. It seems
that the proto-oncogene may thus be brought under the control of the
immunoglobulin promotor, which
is presumably very active in B lymphocytes. This explains why this tumor arises in B
cells. In other lymphomas, a c-onc is brought next to the immunoglobulin light chain
promotor. These are also B cell lymphomas.
Epstein-Barr virus is probably the cause of Burkitt's lymphoma. This is
a herpes virus and herpes viruses
commonly cause chromosomal breaks. If such a break causes an 8:14 translocation,
the cell's myc gene will become adjacent to the cell's immunoglobulin promotor
and c-myc expression will rise in the cells in which this promotor is active.
|
| |
Is there evidence that
mutations in cellular oncogenes might also result in transformation?
The best evidence comes from the cellular oncogene that is the homologue of
the viral oncogene
found in the Harvey strain of murine sarcoma virus (the v-onc is called
HaRas). This c-onc was isolated from bladder carcinomas and compared to the normal
c-onc proto-oncogene. In many tumor cells only one change was found in the amino
acid sequence of the protein; glycine at amino acid position 12 was changed to valine. At position 12 only glycine and proline gave normal
growth. All other amino acids at this position gave a transformed cell. In a lung
carcinoma, the transforming DNA also contained c-HaRas, again it had a point mutation,
this time at position 61.
|
| |
What is the normal function
of oncogenes?
As mentioned above, c-oncs are normal cellular genes that are expressed
and function at some stage of the life of the cell. We should expect them to be
involved in DNA synthesis or perhaps the signaling pathways that lead to proliferation.
More than 40 oncogenes have been identified and there are probably a few
undiscovered ones.
We can sub-divide the cellular oncogenes into those that encode nuclear
proteins and those that encode extra-nuclear proteins. The latter are mostly associated
with the plasma membrane of the cell (Figure 22 and 23).
-
Products of oncogenes that are nuclear proteins: e.g.
myc,
myb.
These are involved in control of gene expression (that is the regulation of
transcription - they are transcription factors) or the control of DNA replication. Neoplasia
is associated with elevated transcription of the oncogene but strong expression is not
always necessary, rather there is a need to make the gene constitutively active rather than
under control of normal regulatory processes.
-
Products of oncogenes that are cytoplasmic or membrane-associated proteins:
e.g. abl, src, ras.
This type does not exhibit altered expression but seems to
convert from proto-oncogene to oncogene by mutation. Thus, in src-induced tumors, strong
over expression of the oncogene has no effect.
|
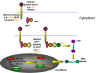 Figure 22
Figure 22
Ways in which altered proto- oncogenes might lead to cell
transformation
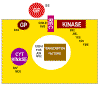 Figure 23
Figure 23
Classes of cellular proto-oncogene products
GF = growth factors
REC = membrane receptors
GP = G-protein transducers of signals
KINASE = membrane bound tyrosine kinase
CYT KINASE = cytoplasmic protein kinase
|
|
FUNCTION OF PROTO-ONCOGENE- ENCODED PROTEINS |
EXAMPLE |
|
Control of DNA transcription (found in nucleus) |
myc |
|
Signaling of hormone/growth factor binding such as a tyrosine
kinase |
src
is a membrane-bound tyr kinase. |
| GTP-binding proteins involved in
signal transduction from a surface receptor to the nucleus |
ras |
|
Growth factors |
sis is an altered form of
platelet-derived growth factor B chain |
|
Growth factor receptors |
erb-B is a homolog of the
epidermal growth factor receptor (it is also a tyrosine kinase). fms is a
homolog of the macrophage colony-stimulating factor (M-CSF) receptor |
In each of these cases, the mutation is dominant.
Thus, for example if
one allele of erb-B (a homolog of the EGF receptor) is mutated so that it is a constitutively switched on (i.e. does not
need epidermal growth factor to bind to switch on the tyrosine kinase activity), then the
signal is on, regardless of the fact that the other allele is normal.
|
 Figure 24A Figure 24A
Dominant mutations are function
gained
|
ANTI-ONCOGENES (Tumor suppressor genes)
The way in which retroviruses cause tumor formation via oncogenes was
established before anything was known about how DNA tumor viruses cause tumors. Certainly,
DNA tumor viruses carry oncogenes (e.g. SV40 T-antigen) but how do these proteins, encoded
in true viral genes with no cellular homologs, cause the formation of tumors?
It has long been known that most tumors are the result of dominant
mutations, i.e. a function is gained that makes the cell grow when it should
not
(Figure 24). For example, as noted above, if we have a receptor that sends a signal when it binds a growth factor
by switching on its tyrosine kinase activity and that receptor becomes mutated so that its
tyrosine kinase activity is permanently activated, the cell will get the aberrant growth
signal even in the heterozygote. Thus the mutant allele is dominant over the normal
allele.
|
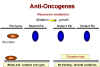 Figure 24B
Figure 24B
Recessive mutations are functions lost
|
Retinoblastoma: A recessive tumor
There is a curious class of tumors that do not fit the usual
characteristics in which the mutant oncogene is dominant over the wild type.
In retinoblastoma, there appears to be a lesion that is recessive, that is
the cancer causing mutation causes a loss of function
(Figure 24). (This is recessive
because, in a diploid organism, there are two genes. If one allele is mutated so that it
does not work, the other can still code for the normal protein and function is retained.
In order to lose the function and have no protein being made, both genes must be
mutated, i.e. we have a recessive mutation). Thus it appears that the protein that is
encoded in the retinoblastoma (Rb) gene is a growth suppressor. If a homozygous
mutation occurs in the Rb gene, there will be no Rb gene product at all and the cell will
grow abnormally because the growth suppressor is no longer present. The product of the Rb
gene has been identified and shown to be a nucleus-located protein of 105 kDaltons.
A heterozygote at the Rb allele still has normal Rb and tumors can still be suppressed
but homozygote has no functional Rb and tumors cannot be suppressed
|
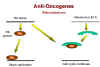 Figure 25
Figure 25
Rb and adenovirus
E1A
|
Above, we have noted that the adenovirus E1A (early function)
protein is somehow involved in tumorigenesis. It has been found that E1A protein in the
transformed adenovirus-infected cell is complexed with a 105kD protein! This turns out to
be the Rb gene product (Figure 25). Thus, it seems that adenovirus may cause a cell to grow abnormally
by complexing (and thereby inactivating) a cellular protein whose normal function is
growth inhibition.
|
| |
p53 and human cancer
Over the past two decades, since its discovery in 1979, a gene known as
the p53 gene (after the size of its encoded protein) has been linked to many
cancers including many that are inherited. In these inherited cancers, it turns
out that the p53 gene is mutated. Alterations in this protein seem to be the basis (direct or
indirect) of most human cancers. In total, 60% of human cancers involve p53. 80% of colon cancers involve the p53 gene
|
Human cancers that involve p53 |
|
cervix |
liver |
|
breast |
lung |
|
bladder |
skin |
|
prostate |
colon |
|
| |
Initially, it was thought that the p53 gene product caused cancers but
further investigation showed the opposite; p53 is, like the retinoblastoma gene product,
a
tumor suppressor. p53 protein has been referred to as The Guardian of the
Genome since it regulates multiple components of the DNA damage control system.
How does p53 work in a functional cell? Normally, there are only a few of
the suppressor p53 molecules in a healthy cell and these are constantly turning over; but
when the DNA becomes damaged (perhaps by radiation or chemical mutagens) and DNA
replication results, p53 turnover ceases. The rise in p53 stops DNA replication.
p53 is a transcription factor. When it builds up, p53 binds to a
specific site(s) on the chromosomes and switches on other genes and these, in turn, shut
down mitosis. p53 can also act in another way: When it builds up it can set the cell on
course to apoptosis. Whether or not p53 causes reversible growth arrest or
apoptosis depends on the state of cellular activation; for example, extensive, unrepaired
DNA damage can lead to sustained p53 production committing the cell to apoptosis. In inherited cancers, there is a mutation in the p53 gene; often it is
a single point mutation and the protein can no longer bind to its correct site on the DNA
and so cannot suppress DNA replication.
Like the Rb gene, product, you would expect the effects of p53 to be
recessive since the second normal p53 allele should make functional protein and
should shut off DNA replication as usual; however, if you are heterozygous for the
mutation, you are, of course, only one mutation away from carcinogenesis. So why do cells that are heterozygous for the p53 mutation also have
problems? Unfortunately, p53 protein forms tetramers in a ribbon-like array and so if half
of the p53 proteins are mutant, there is a good chance that each tetramer will have one
mutant p53 molecule and this inactivates the tetramer, a dominant-negative effect.
|
 Figure 26
Figure 26
p53, hepatitis C
and papilloma virus
|
Although we have learned a lot from families that inherit p53 mutations,
it is clear that most p53 mutations come from non-inherited environmental factors:
carcinogens (benzopyrene in smoke, aflatoxin in molds on peanuts and corn, UV light) that
result in point mutations. There are also gain of function p53
mutations that lead to very aggressive tumors. These turn on DNA replication genes.
What has this got to do with DNA tumor viruses? Just as with
retinoblastoma gene product, the presence of a virus can mimic mutation and take the tumor
suppressor out of action by complexing it in an inactive form that cannot bind to the
specific site on DNA. This is what appears to happen in hepatitis C which
causes hepatocellular
carcinoma. In the case of a human papilloma virus-infected cell, p53
is bound by the E6 protein and directed to a protease that recognizes a cleavage site in p53, thereby destroying
it (Figure 26). In addition, E7 protein binds and inactivates Rb protein.
Much research is now going on to see whether one can
introduce healthy p53 genes into cells to shut down tumor growth.
Thus, our knowledge of how retroviruses cause cancer has led to an
understanding of the formerly cryptic manner in which DNA tumor viruses do the same thing.
|
|


|
 Return to the Virology Section of Microbiology and Immunology On-line
Return to the Virology Section of Microbiology and Immunology On-line
 Return to the
front page of Microbiology and Immunology On-line Return to the
front page of Microbiology and Immunology On-line
This page last changed on
Sunday, June 05, 2016
Page maintained by
Richard Hunt
|

 Figure 12
Figure 12 Figure 13
Figure 13 Figure 14
Figure 14  Figure 16
Figure 16  Figure 17
Figure 17 Figure 18
Figure 18 Figure 19
Figure 19
 Figure 22
Figure 22 Figure 24A
Figure 24A Figure 24B
Figure 24B Figure 25
Figure 25 Figure 26
Figure 26






