Dr. Margaret Hunt
MEDICAL MICROBIOLOGY
MBIM 650/720
READING: Murray et al., 3rd Ed., Chapter 6, appropriate parts of Chapters 50-52

VIROLOGY - CHAPTER THREE
DNA VIRUS REPLICATION STRATEGIES
TEACHING OBJECTIVES
Descriptive analysis of the replicative strategies employed by animal DNA viruses
Identification of virus prototypes associated with different DNA virus replication schemes
GENERAL
Viral genomes contain information which:
ensures replication of viral genomes
ensures packaging of genomes into virions
alters the structure and/or function of the host cell to a greater or lesser degree
VIRAL STRATEGY
Viral strategy refers to the manner in which each virus carries out the above functions. Since a virus is an intracellular parasite, it has to operate within limits imposed by the host cell, or circumvent these limitations.
DNA VIRUS REPLICATION STRATEGIES
General
The virus needs to make mRNAs that can be translated into protein by the host cell translation machinery.
The virus needs to replicate its genome.
Host enzymes for mRNA synthesis and DNA replication are nuclear (except for those in mitochondrion) and so, if a virus is to avail itself of these enzymes, it needs to enter the nucleus.
NUCLEAR DNA VIRUSES
PAPOVAVIRUSES
(The name papovavirus come from papilloma, polyoma, simian vacuolating virus 40)
|
Properties of Papovaviruses |
| They are small: 40-60nm |
| They are icosahedral: major capsid protein is VP1, with lesser amounts of VP2, VP3 |
| They are non-enveloped |
| They have circular, double-stranded DNA is associated with cell histones (nucleosomes) |
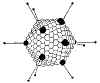
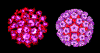 Figure 1 Papilloma virus © Dr Linda
Stannard, University of
Cape Town, South Africa. Used with permission
Figure 1 Papilloma virus © Dr Linda
Stannard, University of
Cape Town, South Africa. Used with permissionThis family contains two genera: PAPILLOMAVIRUSES AND POLYOMAVIRUSES.
PAPILLOMAVIRUSES
Papilloma viruses (figure 1) are difficult to grow in culture. They have a different replication strategy from the polyomaviruses. Papillomaviruses will not be discussed further in this section (but see section of DNA tumor viruses).
 Figure 2 SV40 virus, a polyoma virus © Dr J-Y
Sgro, University of Wisconsin. Used with permission
Figure 2 SV40 virus, a polyoma virus © Dr J-Y
Sgro, University of Wisconsin. Used with permissionPOLYOMAVIRUSES
These include SV40, BK, JC and polyoma viruses. All have a similar strategy for DNA replication. Depending on the host cell, they can either transform the cell (go here) or replicate the virus and lyze the cell.
LYTIC CYCLE
ATTACHMENT, PENETRATION AND UNCOATING
Viral capsid proteins interact with cell surface receptors and penetration is probably via endocytosis. Virions are transported to the nucleus and uncoated. DNA (and associated histones) enters nucleus, probably through a nuclear pore
PRODUCTION OF VIRAL mRNAS AND PROTEINS
Gene expression is divided into early and late phases.
Generalization:
Early genes encode enzymes and regulatory proteins needed to start viral replication processes.
Late genes encode structural proteins, proteins needed for assembly of the mature virus.
Note: - - - - indicates regions of the primary transcript which are removed in the alternatively processed mRNA. Modified from Fiers et al.,Nature 273:113
 Figure 4 Late gene expression
Figure 4 Late gene expression
Note: - - - -
indicates regions of the primary transcript which are removed in the
alternatively processed mRNA.
Modified from Fiers et al.,Nature 273:113
EARLY PHASE OF THE LYTIC CYCLE
Early gene expression (figure 3)
The early promoter is recognized by host RNA polymerase II (SV40 contains a strong enhancer). Post transcriptional RNA modification (capping etc.) is carried out by host enzymes. The early transcript (primary transcript) is made and then undergoes alternative processing, resulting in the mRNAs for the small T and large T antigens (these proteins have common amino-termini but different carboxy-termini).
The mRNAs are translated in the cytoplasm.
Note: Primary transcripts which can be processed and code for more than one protein are seen in several virus families and in the host cell.
LATE PHASE OF THE LYTIC CYCLE
By definition the late phase starts with the onset of viral genome replication.
DNA replication
SV40/polyoma DNA replication occurs in the nucleus:
Large T antigen is needed for DNA replication. This binds to the origin of replication.
DNA replication uses host cell DNA polymerase, which recognizes the viral origin of replication if the T antigen is present.
Replication is bidirectional (There are two replication forks per circular DNA genome and replication involves leading/lagging strands, Okazaki fragments, DNA ligase, etc.). This process of DNA replication is very similar to that which occurs in the host cell - which is not surprising as the virus is using mainly host machinery except for the involvement of the T antigen.
Host histones complex with the newly made DNA.
Late gene expression (figure 4)
Late mRNAs are made after DNA replication (a lot of newly made viral DNA is now available as template). Early mRNAs are still transcribed, but at a much lower rate.
T antigen is involved in controlling the switch on of late transcription and decreased transcription from early promoter.
VP1, 2, and 3 are made from same primary transcript which undergoes differential splicing (figure 5). This results in the reading frame for VP1 being different from that for VP2 and VP3
ASSEMBLY
VP1, 2 and 3 mRNAs are translated in the cytoplasm, the proteins are transported to nucleus, and capsids assemble with DNA (and cell histones) inside capsid.
Capsid particles are released by cell lysis.
 Figure 5 VP1, 2, and 3 are made from same primary transcript which
undergoes differential splicing. This results in the reading frame for VP1
being different from that for VP2 and VP3
Figure 5 VP1, 2, and 3 are made from same primary transcript which
undergoes differential splicing. This results in the reading frame for VP1
being different from that for VP2 and VP3
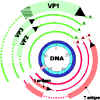 Figure 6
Figure 6Gene expression by SV40. Early genes are in red, late genes are in green. Note: - - - - indicates regions of the primary transcript which are removed in the alternatively processed mRNA. Cross-hatched area indicates region of RNA translated in different reading frames according to which alternatively spliced transcript is being translated Modified from Fiers et al.,Nature 273:113
|
FEATURES TO NOTE ABOUT POLYOMA VIRUS STRATEGY |
| Early and late functions |
| Multiple use of the same DNA sequence (alternative splicing, overlapping reading frames) |
| Multifunctional protein (T antigen) |
| Small genome - so not surprising that virus codes for a very limited number of proteins |
| Host cell provides RNA synthesis machinery, RNA modification machinery, DNA synthesis machinery, histones for packaging DNA. |
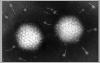 Figure 7a Adenovirus structure © Dr Linda
Stannard, University of Cape
Town, South Africa. Used with permission
Figure 7a Adenovirus structure © Dr Linda
Stannard, University of Cape
Town, South Africa. Used with permission
ADENOVIRUSES
|
PROPERTIES OF ADENOVIRUSES |
| Larger than papovaviruses (70nm diameter) |
| Non-enveloped, icosahedral viruses with fibers (figure 7 and 8) |
Genome 3-5 times size of papovavirus genome |
| The DNA is linear, double stranded, associated with virally coded, basic proteins in virion (unlike papovaviruses, does not use cell histones to package virion DNA) |

B
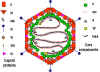 Figure 8
Models of the adenovirus virion. A: A 3-dimensional
image reconstruction of the intact adenovirus particle viewed along an
icosahedral 3-fold axis (© EMBL Virus Structure Resource). B: A stylized section of the adenovirus particle
based on current understanding of its polypeptide components and DNA. No real
section of the icosahedral virion would contain all the components. Virion
constituents are designated by their polypeptide numbers with the exception of
the terminal protein (TP).
Adapted from Fields et al., Fundamental Virology (1996).
Figure 8
Models of the adenovirus virion. A: A 3-dimensional
image reconstruction of the intact adenovirus particle viewed along an
icosahedral 3-fold axis (© EMBL Virus Structure Resource). B: A stylized section of the adenovirus particle
based on current understanding of its polypeptide components and DNA. No real
section of the icosahedral virion would contain all the components. Virion
constituents are designated by their polypeptide numbers with the exception of
the terminal protein (TP).
Adapted from Fields et al., Fundamental Virology (1996).LYTIC CYCLE
ADSORPTION AND PENETRATION
Adenoviruses usually infect epithelial cells. The fibers bind to a cell surface receptor and the virus is engulfed by a clathrin-coated pit. Uncoating occurs in steps. DNA is released into the nucleus (probably at a nuclear pore) (figure 9).
EARLY PHASE
Early transcription:
Adenovirus uses host cell RNA polymerase.
Early mRNAs are transcribed from scattered regions of both strands (figure 10).
Multiple promoters result in more flexible control.
mRNAs are processed by host cell capping, methylation, polyadenylation and (sometimes) splicing enzyme systems, they are then exported to the cytoplasm and translated.
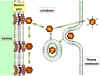 Figure
9 Diagrammatic
representation of the uptake and uncoating of
adenovirus particles.
Adapted from Zinsser Microbiology 20th
Ed.
Figure
9 Diagrammatic
representation of the uptake and uncoating of
adenovirus particles.
Adapted from Zinsser Microbiology 20th
Ed.
 Figure 10
Figure 10
Transcription map of adenovirus. Early genes are shown in
red. Black indicates late genes. Blue lines indicate DNA. Square brackets
indicate the promotors. Missing regions indicate removal of introns. Adapted
from Broker, T.R. In Processing of RNA. (Apirion, D ed) 181-212, 1984
The early proteins include those which:
are needed for transcription (E1A protein is needed for transcription of the other early genes)
are needed for adenovirus DNA synthesis (includes DNA polymerase)
alter expression of host cell genes.
This includes genes whose products:
interfere with the host anti-viral response
interfere with cell cycle regulation
 Figure 11 Adenovirus DNA replication by a
displacement mechanism
Figure 11 Adenovirus DNA replication by a
displacement mechanismLATE PHASE
DNA replication:
Adenovirus encodes its own DNA polymerase (which is one of the early proteins). The DNA is replicated by a strand displacement mechanism (figure 11). There are no Okazaki fragments, both strands are synthesized in a continuous fashion.
DNA polymerases cannot initiate synthesis de novo, they need a primer. In the case of adenovirus, the virally coded terminal protein (TP) (not shown in diagram) acts as a primer. It is thus found covalently linked to the 5' end of all adenovirus DNA strands.
 Figure 12
Figure 12Transcription map of adenovirus. Early genes are shown in red. Green indicates late genes. Blue lines indicate DNA. Square brackets indicate the promotors. Missing regions indicate removal of introns. Adapted from Broker, T.R. In Processing of RNA. (Apirion, D ed) 181-212, 1984
Late transcription:
The way in which late transcription is switched on is not well understood. Late mRNAs code predominantly for structural proteins and there is ONE major late promoter (figure 12). The primary transcript is processed to generate various monocistronic mRNAs (figure 12 and 13):
There are two types of cleavage of primary transcript:
i. to generate various 3' ends which are then polyadenylated
ii. for intron removal
It is not understood how this process is controlled such that the correct amounts of each mRNA are made.
ASSEMBLY
Assembly of adenovirus particles occurs in the nucleus. DNA enters the particles after immature capsids are formed. The capsids then undergo a maturation process, after which the cells lyse and virions leak out.
More structural proteins are made than are needed and excess structural proteins accumulate in the nucleus where they form inclusion bodies.
| FEATURES TO NOTE ABOUT ADENOVIRUS STRATEGY |
| Adenoviruses are larger and more complex than papovaviruses. |
| Adenoviruses code for their own DNA polymerase and DNA packaging proteins. |
| However, although adenoviruses code for their own DNA polymerase, they use host factors in addition to viral proteins for DNA replication, and they use host RNA polymerase and RNA modification systems and so nucleic acid synthesis needs to be in the nucleus. |
 Figure
14a Herpes virus structure
Figure
14a Herpes virus structure
 Figure 14b Herpes simplex virus © Dr Linda M
Stannard,
University of Cape Town, South Africa, 1995 (used with permission).
Figure 14b Herpes simplex virus © Dr Linda M
Stannard,
University of Cape Town, South Africa, 1995 (used with permission).
HERPESVIRUSES
|
PROPERTIES OF HERPES VIRUSES |
| Larger virions (180-200nm) than adenoviruses |
| Larger genome (90-145x106) than adenoviruses |
| Linear, double-stranded DNA |
| Enveloped icosahedral virus (this means that lipid solvents readily inactivate these viruses) (figure 14) |
 Figure 15a Herpes simplex virus adsorbing to the plasma membrane ©
Dennis Kunkel Microscopy, Inc.
Used with permission
Figure 15a Herpes simplex virus adsorbing to the plasma membrane ©
Dennis Kunkel Microscopy, Inc.
Used with permission
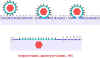 Figure 15b Fusion of membrane-bound virus with
the plasma membrane
Figure 15b Fusion of membrane-bound virus with
the plasma membrane
ADSORPTION AND PENETRATION
Some herpesviruses, including herpes simplex virus, can fuse directly with the cell plasma membrane (results in partial uncoating) (figure 15).
Implications of fusion with plasma membrane:
i) Since the fusion protein is active at physiological pH, if it is inserted into the host cell membrane during the virus growth cycle, that cell can potentially fuse with other cells and form syncytia.
ii) In addition, viral membrane leaves "footprint" - possible clue that the cell is infected (figure 15b)
Capsids are transported towards the nucleus and the DNA passes into the nucleus (probably via nuclear pores).
 Figure
16 Expression of immediate early, early and late genes of herpesviruses
Figure
16 Expression of immediate early, early and late genes of herpesvirusesEARLY PHASE
Early transcription (the mRNAs made during this phase are the alpha and beta mRNAs) (figure 16)
Early transcription uses host RNA polymerase. (A virion protein enters the nucleus upon infection and is important as part of the transcription factor complex recognized by the host RNA polymerase.) The virus uses host mRNA modification enzymes.
Initially, α-mRNAs are transcribed. These are exported to the cytoplasm and translated into alpha-proteins. The α-proteins translated in the cytoplasm are transported into nucleus.
The α-proteins enable the ß-promoters to be used by the host RNA polymerase (figure 16).
ß-mRNAs are transcribed the by host RNA polymerase.
(ß-genes are still "early" since they are transcribed prior to DNA synthesis. Sometimes therefore, α-genes are called "immediate early" and ß-genes are called "early"). ß-proteins are involved in gene expression regulation (they decrease α-gene expression and are needed for γ-gene expression) and in various aspects of DNA synthesis: e.g. herpes β-genes code for:
DNA polymerase
DNA binding proteins
thymidine kinase
ribonucleotide reductase
SINCE THESE BETA PROTEINS ARE VIRALLY-CODED AND NOT HOST-CODED ENZYMES, THEY ARE POTENTIALLY WEAK LINKS IN THE VIRUS LIFE CYCLE AND THUS PROMISING TARGETS FOR VIRAL CHEMOTHERAPY
 Figure 17 Rolling circle DNA replication
Figure 17 Rolling circle DNA replication
LATE PHASE
DNA replication:
DNA circularizes in cell, replicates via a rolling circle mechanism (figure 17), forming tandem repeats which are then cleaved. A simplified scheme is shown at the left.
Some herpesviruses have a complicated genome structure in which two parts of the genome can invert relative to each other, e.g. herpes simplex virus, others do not (figure 18). The significance of this is unclear.
Late transcription:
Late transcription occurs after DNA replication (by definition). γ-mRNAs are made, these are translated in the cytoplasm. γ-proteins are predominantly structural.
There is decreased expression of ß-genes in the late stage. This is probably due to down-regulation of transcription of ß-genes by one of the γ-proteins.
In herpesviruses there is no apparent organization of the genome into blocks for either early or late transcription.
 Figure 19 Stages in the exocytosis of herpes virus from the nucleus, in which the virus
core is assembled, to the plasma membrane
Figure 19 Stages in the exocytosis of herpes virus from the nucleus, in which the virus
core is assembled, to the plasma membraneASSEMBLY
Assembly occurs in the nucleus. A capsid is formed and the DNA enters the capsid. The capsids bud through localized areas of the inner nuclear membrane which have viral membrane proteins inserted into them (figure 19). (These areas have tegument proteins associated with the inner face of the inner nuclear membrane). In some undefined way, virions are released to the environment. Figure 19 indicates one possible route.
Excess structural proteins accumulate in nucleus, often form inclusion bodies (part of the cytopathic effect).
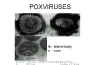 Figure 20 Negative stain and thin section of pox viruses © F.
Fenner
Figure 20 Negative stain and thin section of pox viruses © F.
Fenner
CYTOPLASMIC DNA VIRUSES
POXVIRUSES
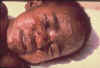 Figure 21 Boy with smallpox CDC/Cheryl Tryon
ctt1@cdc.gov
Figure 21 Boy with smallpox CDC/Cheryl Tryon
ctt1@cdc.gov There are several reasons why poxviruses (figure 20) are of importance:
Certain poxviruses are of historic note such as smallpox (figure 21) and vaccinia (cow pox, which was used in the smallpox vaccine (go here))
- Pox viruses are used in new techniques of vaccine development (such as genetically-engineered vaccinia)
- Some members of this family infect man (molluscum contagium (figure 22), orf, monkey pox)
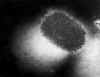 Figure 22 Transmission electron micrograph of poxvirus of molluscum contagiosum
CDC
Figure 22 Transmission electron micrograph of poxvirus of molluscum contagiosum
CDC |
PROPERTIES OF POXVIRUSES |
| large virions |
| large genome, double-stranded DNA 130 - 240x106 mol. wt. |
| complex morphology |
| enveloped |
Poxviruses replicate in the cytoplasm. This means that they must provide their own mRNA and DNA synthetic machinery.
Vaccinia is the most intensively studied member of the poxvirus family.
ADSORPTION AND PENETRATION
The virus binds to cell surface receptors. It enters cells via clathrin-coated pits or by direct fusion of the virus with the plasma membrane. The virus is then released into the cytoplasm, minus its membrane.
EARLY PHASE
Early transcription
After the initial phase of uncoating has occurred, the virus can make a limited number of mRNAs (the immediate early mRNAs). To do this, the poxvirus needs a DNA-dependent RNA polymerase. Host RNA polymerase is in the cell nucleus and so this explains why poxviruses use a virally-coded DNA-dependent RNA polymerase to make their RNAs. Since this enzyme is needed immediately upon infection, it must be brought into the infected cell with the vaccinia DNA, it is thus present in virions.
("Naked" vaccinia DNA which has had all the protein removed is thus not infectious, since it will have no RNA polymerase associated with it, and nothing can happen in the vaccinia life cycle without the vaccinia RNA polymerase.)
Poxvirus mRNAs are capped, methylated and polyadenylated just like standard eucaryotic mRNAs, but host cell mRNAs are modified in the nucleus and vaccinia replicates in the cytoplasm. Since Vaccinia is cytoplasmic, these modifications must be carried out by virally-coded enzymes. The modifying enzymes are packaged in virions and thus those mRNAs made immediately upon infection can be modified. So far, no spliced mRNAs have been reported for vaccinia (this is not surprising since it replicates in cytoplasm and host splicing enzymes are in the nucleus).
One of the immediate early mRNA translation products is an uncoating enzyme. This allows further uncoating of the vaccinia DNA and more genes can now be transcribed - the early genes are now all expressed. Poxviruses are exceptional in that they code for an uncoating protein which has to be made in the newly infected cell before uncoating can be completed.
Virus production "factories" are seen in the cytoplasm - sites of vaccinia virus replication.
The early proteins are involved in DNA replication, RNA transcription, RNA modification and uncoating. They also include a few structural proteins.
LATE PHASE
DNA synthesis
DNA synthesis occurs in "factories" and uses an unusual mechanism which will not be dealt with here.
Late transcription and translation
This is a complex process:
Some late proteins are made throughout the late phase, but others only at the beginning of late phase. Some early proteins are not synthesized once DNA replication commences while other early proteins are made during late as well as early phases. Thus, there is complex control of which proteins are made by vaccinia and when they are made. This means that there are controls other than just early/late controls. (This is a very large virus, thus the complexity is not surprising.)
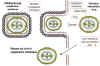 Figure 23 Possible scheme for the formation of infectious pox virions.
The virus core becomes wrapped in cytoplasmic membrane and may escape when the
host cell is lyzed. Some other membrane-bound virions may bud through other
membranes, in which case they have two membranes. In either case, the virions are
infectious.
Adapted from Baron, S. Ed. Medical Microbiology 4th Edition.
1996.
Figure 23 Possible scheme for the formation of infectious pox virions.
The virus core becomes wrapped in cytoplasmic membrane and may escape when the
host cell is lyzed. Some other membrane-bound virions may bud through other
membranes, in which case they have two membranes. In either case, the virions are
infectious.
Adapted from Baron, S. Ed. Medical Microbiology 4th Edition.
1996. ASSEMBLY
Assembly occurs in "factories" in the cytoplasm. The new, immature virus particles acquire a membrane while in the cytoplasm - the exact mechanism is not fully understood but it seems that the virus gets "wrapped" by cellular membranes (figure 23). There is a gradual maturation of enveloped particles. The virus is usually released by host cell disintegration, but some may get out by budding through membranes (in which case they have an extra membrane). Both forms appear to be infectious.
The exact mechanism by which the virus gets out of infected cells may depend on host cell type.
![]() Return to the Virology section of Microbiology and Immunology On-line
Return to the Virology section of Microbiology and Immunology On-line
![]() Return to the Department of Microbiology and Immunology Site Guide
Return to the Department of Microbiology and Immunology Site Guide
This page
copyright 2003, The Board of Trustees of the University of South Carolina
This page last changed on
Tuesday, October 19, 2004
Page maintained by Richard Hunt
URL: http://www.med.sc.edu:85/mhunt/dna1.htm
Please report any problems to rhunt@med.sc.edu