| x | x | ||||
 |
|||||
| BACTERIOLOGY | IMMUNOLOGY | MYCOLOGY | PARASITOLOGY | VIROLOGY | |
|
|
|
||||
| TURKISH | |||||
| PORTUGUESE | |||||
| SPANISH | |||||
| SHQIP - ALBANIAN | |||||
| FARSI | |||||
|
|
|||||
|
Let us know what you think |
|||||
| SEARCH | |||||
|
|
|||||
| Logo image © Jeffrey Nelson, Rush University, Chicago, Illinois and The MicrobeLibrary | |||||
|
|
|||||
|
KEY WORDS Isolation (culture)
Identification & taxonomy Biochemical (physiological) tests Molecular tests Chemical profiling Non culture based
detection |
Bacterial identification in the diagnostic laboratory versus taxonomy Isolation and identification of bacteria from patients aids treatment since infectious diseases caused by different bacteria have a variety of clinical courses and consequences. Susceptibility testing of isolates (i.e. establishing the minimal inhibitory concentration or MIC) can help in selection of antibiotics for therapy. Recognizing that certain species (or strains) are being isolated atypically may suggest that a disease outbreak has occurred e.g. from contaminated hospital supplies or poor aseptic technique on the part of hospital personnel. When patients are suspected of having a bacterial infection, it is usual to isolate visible colonies of the organism in pure culture (on agar plates), and then speciate the organism. The identification is based on taxonomic principles applied to the clinical microbiological situation. In the diagnostic laboratory, many samples must be characterized each day and results obtained as quickly as possible. Tests must be easily learned, low in cost and rapidly performed. These classical methods for speciation of bacteria are based on morphological and metabolic characteristics. The diagnostic tests have been selected on the basis that empirically they provide discriminating information. There are numerous different tests for each of the many target pathogens. Additionally, molecular biology techniques (for characterization of specific genes or gene segments) are now commonplace in the clinical laboratory. Modern taxonomic approaches often employ technically more complex methodology and are concerned with profiling the structural composition of bacteria. This often involves "molecular biology" or "analytical chemistry" -based approaches. It is now recognized that many of the classical schemes for differentiation of bacteria provide little insight into their genetic relationships and in some instances are scientifically incorrect. New information has resulted in renaming of certain bacterial species and in some instances has required totally reorganizing relationships within and between many bacterial families. |
||||
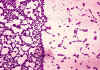
 Figure 1 Bicarbonate and blood agar plate cultures of
Bacillus anthracis. Smooth colonies on bicarbonate
(left) and rough colonies on blood agar (right). CDC/Dr. James Feeley
Figure 1 Bicarbonate and blood agar plate cultures of
Bacillus anthracis. Smooth colonies on bicarbonate
(left) and rough colonies on blood agar (right). CDC/Dr. James Feeley
|
Taxonomic terms (classification) Family: a group of related genera. Genus: a group of related species. Species: a group of related strains. Type: sets of strain within a species (e.g. biotypes, serotypes). Strain: one line or a single isolate of a particular species. The most commonly used term is the species name (e.g. Streptococcus pyogenes - abbreviation S.pyogenes). There are always two parts to the species name, one defining the genus in this case "Streptococcus" and the other the species (in this case "pyogenes"). The genus name is always capitalized but the species name is not. Both species and genus are underlined or in italics.
Steps in diagnostic isolation and identification of bacteria Step 1. Samples of body fluids (e.g. blood, urine, cerebrospinal fluid) are streaked on culture plates and isolated colonies of bacteria (which are visible to the naked eye) appear after incubation for one to several days (Figure 1). Each colony consists of millions of bacterial cells. Observation of these colonies for size, texture, color, and (if grown on blood agar) hemolysis reactions, is highly important as a first step in bacterial identification. Whether the organism requires oxygen for growth is another important differentiating characteristic. Step 2. Colonies are Gram stained and individual bacterial cells observed under the microscope. Step 3. The bacteria are speciated using these isolated colonies. This often requires an additional 24 hours of growth. |
||||
|
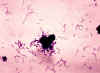
FIGURE 3. EXAMPLES OF GRAM STAINS
|
The Gram Stain
A colony is dried on a slide and treated as follows (Figure 2 and 3): Step 1. Staining with crystal violet. Step 2. Fixation with iodine stabilizes crystal violet staining. All bacteria remain purple or blue. Step 3. Extraction with alcohol or other solvent. Decolorizes some bacteria (Gram negative) and not others (Gram positive). Step 4. Counterstaining with safranin. Gram positive bacteria are already stained with crystal violet and remain purple. Gram negative bacteria are stained pink. Under the microscope, the appearance of bacteria is observed. Questions to be asked include:
Besides the Gram stain, there are other less commonly employed stains available (e.g. for spores and capsules). Another similar colony from the primary isolation plate is then examined for biochemical properties; for example, will the bacteria ferment a sugar such as lactose? In some instances, the bacteria are identified (e.g. by aggregation) with commercially available antibodies recognizing defined surface antigens. Other commercial molecular tests are now widely used.
There is considerable diversity even within a
species. Thus comparisons of species involve comparisons of multiple strains for
each species. Comparisons are primarily based on chemical or molecular analysis. Chemical analysis Molecular analysis DNA-DNA homology (or how well two strands of DNA from different bacteria bind [hybridize] together) is employed to compare the genetic relatedness of bacterial strains/species. If the DNA from two bacterial strains display a high degree of homology (i.e. they bind well), the strains are considered to be members of the same species. DNA from different bacterial species (unless closely related) display no homology. In the last few years, sequencing of 16S ribosomal
RNA molecules (16S rRNA) has become the "gold standard" in bacterial
taxonomy. The molecule is approximately sixteen hundred nucleotides in length.
The sequence of 16S rRNA provides a measure of genomic similarity above the
level of the species allowing comparisons of relatedness across the entire
bacterial kingdom. Closely related bacterial species often have identical rRNA
sequences. The technique thus provides complementary information to DNA-DNA
hybridization. Determinations of the sequence of 16S rRNA genes and other
genetic regions are used in identification in the clinical microbiology
laboratory. Approaches to rapid diagnosis without prior culture Certain human pathogens (including the causative agents of tuberculosis, Lyme disease and syphilis) either cannot be isolated in the laboratory or grow extremely poorly. Successful isolation can be slow and in some instances impossible. Direct detection of bacteria without culture is possible in some cases. A simple approach to rapid diagnosis (as an example of antigen detection) is used in many doctor's offices for the group A streptococcus. The patient's throat is swabbed and streptococcal antigen extracted directly from the swab (without prior bacteriological culture). The bacterial antigen is detected by aggregation (agglutination) of antibody coated latex beads. Bacterial DNA sequences can be amplified directly from human body fluids (the polymerase chain reaction, PCR). In this fashion large amounts of specific genes or portions of genes can be generated and readily detected. For example, great success has been achieved in rapid diagnosis of tuberculosis. Finally, direct microscopic observation of certain clinical samples for the presence of bacteria can be helpful (e.g. detection of M. tuberculosis in sputum). Serologic identification of an antibody response
(in patient's serum) to the infecting agent can only be successful several weeks
after an infection has occurred. |
||||
|
|
|||||
|
|
|||||
|
ANIMATION |
|||||
|
TUTORIAL |
|||||
|
|
|
||||