|
x |
x |
|
 |
 |
|
INFECTIOUS
DISEASE |
BACTERIOLOGY |
IMMUNOLOGY |
MYCOLOGY |
PARASITOLOGY |
VIROLOGY |
|
VIETNAMESE |
IMMUNOLOGY - CHAPTER TEN
MAJOR HISTOCOMPATIBILITY COMPLEX (MHC)
AND T-CELL RECEPTORS - ROLE IN IMMUNE RESPONSES
Gene Mayer, Ph.D
Emertius Professor of Pathology, Microbiology and Immunology
University of South Carolina
Jennifer Nyland, Ph.D
Assistant Professor of Pathology, Microbiology and Immunology
University of South Carolina
|
|
TURKISH |
|
FRANCAIS |
|
PORTUGUESE |
Let us know what you think
FEEDBACK |
|
SEARCH |
|
|

 |
|
Logo image © Jeffrey
Nelson, Rush University, Chicago, Illinois and
The MicrobeLibrary |
|
|
|
|
TEACHING OBJECTIVES
To give an overview of the role of the major histocompatibility complex
in immune responses.
To describe the structure and function of class I and class II MHC
molecules.
To discuss the nature of polymorphisims in class I and class II MHC
molecules.
To describe the structure of the T cell receptor for antigen
To discuss the genetic basis for the generation of diversity in the TCR.
To discuss the role of the CD3 complex and co-stimulatory molecules.
To describe the nature of the immunological synapse.
To discuss the requirements for T cell activation.
|
Historical Overview
Cell-cell interactions of the adaptive immune response
are critically important in protection from pathogens. These interactions
are orchestrated by the immunological synapse whose primary components are
the T cell antigen receptor (TCR) and the Major histocompatibility complex (MHC)
molecule. The major function of the TCR is to recognize antigen in the
correct context of MHC and to transmit an excitatory signal to the interior
of the cell. Since binding of peptide within the MHC is not covalent, there
are many factors while help stabilize the immunological synapse.
Gene products encoded in the MHC were first identified as being important in rejection of
transplanted tissues. Furthermore, genes in the MHC were found to be highly
polymorphic (i.e. in the population there were many different allelic forms
of the genes). Studies with inbred strains of mice showed that genes in the
MHC were also involved in controlling both humoral and cell-mediated immune
responses. For example, some strains of mice could respond to a particular
antigen but other strains could not and these strains differed only in one
or more of the genes in the MHC. Subsequent studies showed that there were
two kinds of molecules encoded by the MHC – Class I molecules and class II
molecules which are recognized by different classes of T cells. Class I molecules were found on all nucleated cells (not red
blood cells) whereas class II molecules were found only on antigen
presenting cells, (APCs) which included dendritic cells, macrophages, B
cells and a few other types (Figure 1).
It was not until the discovery of how the TCR recognizes antigen that the role of MHC genes in immune
responses was understood. The TCR was shown to recognize antigenic peptides
in association with MHC molecules. T cells recognize portions of protein
antigens that are bound non-covalently to MHC gene products. Cytotoxic T
cells (Tc) recognize peptides bound to class I MHC molecules and helper T
cells (Th) recognize peptides bound to class II MHC molecules. The three
dimensional structures of MHC molecules and the TCR have been determined by
X-ray crystallography so that a clear picture of how the TCR, MHC gene
products and antigen interact has emerged.
|
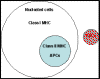 Figure 1
Figure 1
Distribution of class I and class II MHC molecules on human cells
 Figure 2
Figure 2
The MHC class 1 molecule has three globular domains alpha 1 (yellow),
alpha 2 (green) and alpha 3 (blue). The alpha 3 domain is closely
associated with the non-MHC -encoded beta 2 microglobulin (pink). The
latter is stabilized by a disulfide bridge (red) and is similar to an
immunoglobulin domain in three-dimensional structure. The alloantigenic
sites which carry determinants specific to each individual are found in
the alpha 1 and 2 domains. The latter also has a carbohydrate chain
(blue, CHO). There is a phosphate in the cytoplasmic domain. Papain
cleaves near the outer surface of the plasma membrane |
Structure of Class I MHC
Molecules
The molecule
Class I MHC molecules are composed of two polypeptide chains, a
long α chain and a short β chain called β2-microglobulin (figure 2).
The α chain has four regions.
-
A cytoplasmic region, containing
sites for phosphoylation and binding to cytoskeletal elements.
-
A transmembrane region containing hydrophic
amino acids by which the molecule is anchored in the cell
membrane.
-
A highly conserved α3 immunoglubilin-like domain to which CD8 binds.
-
A highly
polymorphic peptide binding region formed from the α1 and α2 domains.
The β2- microglobulin associates with the α chain and helps maintain the
proper conformation of the molecule.
The antigen-binding groove
An analysis of which part of the class I MHC molecules is most
variable demonstrates that variability is most pronounced in the α1 and
α2 domains, which comprise the peptide binding region (Figure 3). The
structure of the peptide binding groove, revealed by X-ray
crystallography, shows that the groove is composed of two α helices
forming a wall on each side and eight β-pleated sheets forming a floor.
The peptide is bound in the groove and the residues that line the groove
make contact with the peptide (Figure 4). These are the residues that
are the most polymorphic. The groove will accommodate peptides of
approximately 8-10 amino acids long. Whether a particular peptide will
bind to the groove will depend on the amino acids that line the groove.
Because class I molecules are polymorphic, different class I molecules
will bind different peptides. Each class I molecule will bind only
certain peptides and will have a set of criteria that a peptide must
have in order to bind to the groove. For example, Figure 5 shows that
one class I molecule will bind peptides that have a leucine (L) as the
carboxy-terminal amino acid and either tyrosine (Y) or phenylalanine (F)
as the 4th amino acid from the carboxy-terminal end. As long
as these two conditions are met a peptide will bind, regardless of what
the other amino acids are. Similarly a different class I molecule will
bind any peptide that has a tyrosine (Y) as the second amino acid from
the amino-terminal end and either a valine (V), isoleucine (I) or
leucine (L) at the carboxy-terminal end (Figure 5). Thus, for every
class I molecule, there are certain amino acids that must be a
particular location in the peptide before it will bind to the MHC
molecule. These sites in the peptide are referred to as the “anchor
sites”. The ends of the peptide are buried within the closed ends
of the class I binding groove while the center bulges out for
presentation to the TCR.
Within the MHC there are 6 genes that encode class I molecules
HLA-A, HLA –B, HLA-C, HLA-E, HLA-F and HLA-G. Among these HLA-A, HLA
–B, and HLA-C are the most important and are most polymorphic. Table 1
shows the degree of polymorphism at each of these loci.
|
 Figure 3 Most variability in amino acids at different positions along the alpha
chain of class I MHC molecules occurs in the alpha 1 and alpha 2
regions. The greatest polymorphism is found for amino acids that line
the wall and floor of the groove that binds the peptides
Figure 3 Most variability in amino acids at different positions along the alpha
chain of class I MHC molecules occurs in the alpha 1 and alpha 2
regions. The greatest polymorphism is found for amino acids that line
the wall and floor of the groove that binds the peptides |
 Figure 4
Figure 4
a. Peptide binding groove of class I MHC molecules.
b. Groove with highlighted highly variable residues. The variable
residues are clustered around the peptide-binding pocket
 Figure 5
Figure 5
Anchor sites in peptides that bind to class I MHC molecules (adapted
from Janeway et al. Immunobiology 6th Edition
|
| |
|
Table 1. Polymorphism of class I MHC
genes |
| Locus |
Number
of alleles
(allotypes) |
| HLA-A |
218 |
| HLA-B |
439 |
| HLA-C |
96 |
| HLA-E, HLA-F
and HLA-G |
Relatively
few alleles |
|
|
CHIME

Chime presentation showing the
regions of variability of MHC I molecules and the interaction of the
alpha chain with other subunits of the MHC I complex and the bound
peptide (requires
Chime plug-in. Get Chime here) |
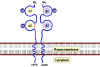 Figure 6 MHC class II molecules comprise two non-identical peptides (alpha and
beta) which are non-covalently associated and traverse the plasma
membrane with the N terminus to the outside of the cell. The
domains closest to the membrane in each chain are structurally
related to immunoglobulins. With the exception of the alpha 1
domain, all domains are stabilized by disulfide bridges (red).
Both the alpha and beta chains are glycosylated. The beta chain is
shorter than the alpha chain (beta mol. wt = 28,000) and contains
the alloantigenic sites. There is some polymorphism in the alpha
chain of some MHC II molecules
Figure 6 MHC class II molecules comprise two non-identical peptides (alpha and
beta) which are non-covalently associated and traverse the plasma
membrane with the N terminus to the outside of the cell. The
domains closest to the membrane in each chain are structurally
related to immunoglobulins. With the exception of the alpha 1
domain, all domains are stabilized by disulfide bridges (red).
Both the alpha and beta chains are glycosylated. The beta chain is
shorter than the alpha chain (beta mol. wt = 28,000) and contains
the alloantigenic sites. There is some polymorphism in the alpha
chain of some MHC II molecules
 Figure 7
Figure 7
The
greatest polymorphism for the beta chain of class II MHC molecules is
found for those amino acids in the beta I region that line the wall
and floor of the groove that binds the peptide |
Structure of Class II MHC Molecules
The molecule
Class II MHC molecules are composed of two polypeptide chains an α and a
β chain of approximately equal length (Figure 6). Both chains have four
regions:
- A cytoplasmic region containing sites for phosphoylation and
binding to cytoskeletal elements
- A transmembrane region
containing hydrophic amino acids by which the molecule is anchored in
the cell membrane
- A highly conserved α2 domain and a highly conserved β2
domain to which CD4 binds
- A highly polymorphic peptide binding region formed from the
α1 and β1 domains
The antigen-binding groove
As with Class I MHC molecules, an analysis of which part of the class II
MHC molecule is most variable demonstrates that variability is most
pronounced in the α1 and β1 domains, which comprise the peptide binding
region (Figure 7). The structure of the peptide binding groove, revealed
by X-ray crystallography, shows that, like class I MHC molecules, the
groove is composed of two α helices forming a wall on each side and
eight β-pleated sheets forming a floor. Both the α1 and β1 chain
contribute to the peptide binding groove. The peptide is bound in the
groove and the residues that line the groove make contact with the
peptide. These are the residues that are the most polymorphic. The
groove of Class II molecules is open at one end so that the groove can
accommodate longer peptides of approximately 13-25 amino acids long with
some of the amino acids located outside of the groove. Whether a
particular peptide will bind to the groove will depend on the amino
acids that line the groove. Because class II molecules are polymorphic,
different class II molecules will bind different peptides. Like class I
molecules, each class II molecule will bind only certain peptides and
will have a set of criteria that a peptide must have in order to bind to
the groove (i.e. “anchor sites”).
Within the MHC there are 5 loci that encode class II molecules, each of
which contains a gene for an α chain and at least one gene for a β
chain. The loci are designated as HLA-DP, HLA –DQ, HLA-DR, HLA-DM, and
HLA-DO. Among these, HLA-DP, HLA –DQ, and HLA-DR are the most important
and are most polymorphic. Table 2 shows the degree of polymorphism at
each of these loci.
|
| |
Important Aspects of MHC
-
Although there is a high degree of polymorphism for a
species, an individual has maximum of six different class I MHC products
and only slightly more class II MHC products (considering only the major
loci).
-
Each MHC molecule has only one binding site. The
different peptides a given MHC molecule can bind all bind to the same
site, but only one at a time.
-
Because each MHC molecule can bind many different peptides,
binding is termed degenerate.
-
MHC polymorphism is determined only in the germline. There
are no recombinational mechanisms for generating diversity.
-
MHC molecules are membrane-bound; recognition by T cells
requires cell-cell contact.
-
Alleles for MHC genes are co-dominant. Each MHC
gene product is expressed on the cell surface of an individual nucleated
cell.
-
A peptide must associate
with a given MHC of that
individual, otherwise no immune response can occur. That is one level
of control.
-
Mature T cells must have
a T cell receptor that recognizes the peptide associated with MHC. This is the second level of control.
-
Cytokines (especially
interferon-γ) increase level of expression of MHC.
-
Peptides from the cytosol associate with class I MHC and
are recognized by Tc cells. Peptides from within vesicles associate
with class II MHC and are recognized by Th cells.
-
Polymorphism in MHC is important for survival of the
species.
|
Table 2. Polymorphism of class II MHC
genes |
| Locus |
Number
of alleles
(allotypes) |
HLA-DPA
HLA-DPB |
12
88 |
HLA-DQA
HLA-DQB |
17
42 |
HLA-DRA
HLA-DRB1
HLA-DRB3
HLA-DRB4
HLA-DRB5 |
2
269
30
7
12 |
| HLA-DM and
HLA-DO |
Relatively
few alleles |
|
| |
How do
peptides get into the MHC groove?
Peptides from the cytosol associate with class I MHC and are
recognized by CTL cells. The peptides enter the endoplasmic
reticulum and bind in the MHC class I groove. This complex is then
exported to the cell surface through the Golgi. MHC class II
molecules are formed with an invariant (Ii) chain as a place holder
while in the ER and Golgi. The Ii chain is cleaved and removed once
the complex is in a vesicle. Peptides from within the vesicle
associate with class II MHC and are then exported to the cell
surface where they are recognized by Th cells.
The role of TCR in the immune
response
The TCR is a surface molecule found on T cells that recognizes
antigen presented in the correct MHC context. The TCR is similar to
immunoglobulin and is part of the immunoglobulin superfamily. There are
two types of TCRs, the predominant αβ which is commonly found in
lymphoid tissues, and the γδ which is found at mucosal surfaces. |
 Figure 8
Figure 8
The T cell receptor heterodimer comprises two transmembrane
glycoproteins, the alpha and beta chains. There are two domains in the
external part of each chain and these resemble immunoglobulin variable
and constant regions. There are sugar chains on each domain.
There is a short sequence similar to the immunoglobulin hinge region
that connects the immunoglobulin-like domains to the transmembrane
sequence. This contains cysteines that form a disulfide bridge. The
hydrophobic transmembrane helical structures are unusual in that they
contain positively charged amino acids (basic amino acids). The alpha
chain has two positively charged residues while the beta chain has
one.
 Structure of A6-T cell receptor bound to MHC
class I molecule complexed with an altered Htlv-1 Tax Peptide Y8a. The HIV peptide is shown in gray. MHC class I molecule is in dark blue,
the associated beta 2 microglobulin in light blue. T cell receptor is
in green and yellow. Y. H.Ding, B. M.Baker, D.
N.Garboczi, W. E.Biddison & D. C.Wiley
MMDB Id: 11766 PDB Id: 1QSF Image prepared using RasMol
Structure of A6-T cell receptor bound to MHC
class I molecule complexed with an altered Htlv-1 Tax Peptide Y8a. The HIV peptide is shown in gray. MHC class I molecule is in dark blue,
the associated beta 2 microglobulin in light blue. T cell receptor is
in green and yellow. Y. H.Ding, B. M.Baker, D.
N.Garboczi, W. E.Biddison & D. C.Wiley
MMDB Id: 11766 PDB Id: 1QSF Image prepared using RasMol
 Figure 9
Figure 9
Rearrangement of the TCR beta chain genes |
Structure of the T cell receptor (TCR)
The TCR is
a heterodimer composed of one α and one β chain of approximately equal
length (Figure 8). Each chain has a short cytoplasmic tail but it is to
small to be able to transduce an activation signal to the cell. Both
chains have a transmembrane region comprised of hydrophobic amino acids
by which the molecule is anchored in the cell membrane. Both chains
have a constant region and a variable region similar to the
immunoglobulin chains. The variable region of both chains contains
hypervariable regions that determine the specificity for antigen. Each
T cell bears a TCR of only one specificity (i.e. there is allelic
exclusion).
THE
GENETIC BASIS FOR RECEPTOR GENERATION
The
genetic basis for the generation of the vast array of antigen receptors
on B cells has been discussed previously (see lecture on Ig genetics).
The generation of a vast array of TCRs is accomplished by similar
mechanism. The germline genes for the TCR β genes are composed of V, D
and J gene segments that rearrange during T cell development to produce
many different TCR β chains (Figure 9). The germline genes for the TCR
α genes are composed of V and J gene segments which rearrange to produce
α chains. The specificity of the TCR is determined by the combination
of α and β chains.
There is a
small population of T cells that express TCRs that have γ and δ chains
instead of α and β chains. These gamma/delta T cells predominate in the
mucosal epithelium and have a repertoire biased toward certain bacterial
and viral antigens. The genes for the δ chains have V, D and J gene
segments whereas the genes for the γ chains have only V and J gene
segments but the repertoire is considerably smaller that than that of
the alpha/beta T cells. The gamma/delta T cells recognize antigen in an
MHC-independent manner unlike the alpha/beta T cells.
Important aspects of the TCR
-
Each T cell bears a TCR of only one
specificity (i.e. there is allelic exclusion).
-
The αβ TCR recognizes antigen only in the
context of cell-cell interaction and in the correct MHC.
-
The γδ TCR recognizes antigen in an MHC-independent
manner in response to certain viral and bacterial antigen.
|
TABLE 3
COMPARISON OF
THE MAJOR PROPERTIES OF IMMUNOGLOBULIN (Ig) AND T-CELL
RECEPTOR (TCR) GENES AND PROTEINS |
|
GENES |
|
Properties |
Ig |
TCR |
|
Many VDJs, Few C's |
Yes |
Yes |
|
VDJ Rearrangement |
Yes |
Yes |
|
V pairs form
antigen-recognition site |
Yes |
Yes |
|
Somatic hypermutation |
Yes |
No |
|
PROTEINS |
|
Transmembrane forms |
Yes |
Yes |
|
Secreted forms |
Yes |
No |
|
Isotypes with distinct
functions |
Yes |
No |
|
Valency |
2 |
1 |
|
Adapted from Janeway and
Travers, Immunobiology |
|
 Structure of a crystal structure of a complex of a human T cell
receptor, influenza Ha Antigen Peptide and an MHC Class II Molecule.
The alpha and beta chains of the MHC II molecules are in dark and
light blue. The T cell receptor is in yellow and green. The influenza
peptide is in gray. Hennecke, J., Carfi, A., Wiley, D.
C. MMDB Id: 14648 PDB Id: 1FYT. Image prepared using RasMol
Structure of a crystal structure of a complex of a human T cell
receptor, influenza Ha Antigen Peptide and an MHC Class II Molecule.
The alpha and beta chains of the MHC II molecules are in dark and
light blue. The T cell receptor is in yellow and green. The influenza
peptide is in gray. Hennecke, J., Carfi, A., Wiley, D.
C. MMDB Id: 14648 PDB Id: 1FYT. Image prepared using RasMol |
CHIME

Click on the image above to view rotatable structure and identify protein chains
of MHC I and TCR interacting with HTLV tax peptide (requires
Chime plug-in. Get Chime
here) |
CHIME

Click on the image above to view a
rotatable structure and identify protein chains of MHC II and TCR
interacting with an influenza HA peptide (requires Chime plug-in. Get Chime
here) |
 Figure 10
Figure 10
The receptor for antigens on the T cell surface comprises eight
proteins.
(a) Two disulfide-bonded chains of the T cell receptor which form a
heterodimer. These recognize peptides associated with MHC molecules.
(b) Four chains, collectively termed CD3, that associate with the T
cell receptor dimer and participate in its transport to the surface of
the cell. The CD3 complex together with the zeta chains, which form a
homodimer, transduce the signal after antigen has bound |
TCR and CD3 Complex
The TCR is closely associated with a group of 5
proteins collectively called the CD3 complex (Figure 10). The CD3
complex is composed of one γ, one δ, two ε and 2 ξ chains. All of the
proteins of the CD3 complex are invariant and they do not contribute to
the specificity in any way. The CD 3 complex is necessary for cell
surface expression of the TCR during T cell development. In addition,
the CD3 complex transduces activation signals to the cell following
antigen interaction with the TCR.
|
 Figure 11
Figure 11
A. Molecules involved in the interaction between T cells and
antigen-presenting cells. Some cytokines produced by each cell type are
shown

B. Ligands involved in the interaction of cytotoxic T cells and
their target cells
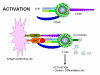 Figure 12a
Figure 12a
Activation of T cells only occurs when both TCR and co-stimulatory
molecules are engaged with their respective ligands
 Figure 12b
Figure 12b
Down regulation occurs if CTLA-4 interacts with B7:
CTLA-4 sends an inhibitory signal
 Figure 12c
Figure 12c
Engagement of TCR and antigen/MHC in the absence of co-stimulation may
lead to anergy
 Figure 12d
Figure 12d
Engagement of co-stimulatory molcules in the absence of TCR engagement
results in no response |
The “Immunological synapse”
The interaction between the TCR and MHC molecules are not very strong. Accessory
molecules are necessary to help stabilize the interaction (Figure 11a,b). These
include:
-
CD4 binding to Class II MCH, which ensures that Th cells only
interact with APCs
-
CD8 binding to class I MHC, which ensures that Tc cells can
interact with target cells
-
CD2binding to LFA-3
-
LFA-1 binding to ICAM-1
The accessory molecules are invariant and do not contribute to the
specificity of the interaction, which is solely determined by the TCR. The
expression of accessory molecules can be increased in response to cytokine,
which is one way that cytokines can modulate immune responses.
In addition to accessory molecules which help stabilize the interaction between
the TCR and antigen in association with MHC molecules, other molecules are also
needed for T cell activation. Two signals are required for T cell activation –
one is the engagement of the TCR with Ag/MHC and the other signal comes from the
engagement of co-stimulatory molecules with their ligands. One of the most
important (but not the only) co-stimulatory molecule is CD28 on T cells which
must interact with B7-1 (CD80) or B7-2 (CD81) on APCs . Like accessory molecules
the co-stimulatory molecules are invariant and do not contribute to the
specificity of the interaction. The multiple interactions of TCR with Ag/MHC and
the accessory and co-stimulatory molecules with their ligands have been termed
the “immunological synapse.”
Not only is co-stimulation necessary for T cell activation, a lack of
co-stimulation may result in anergy (i.e., inability to respond to antigen) or
down-regulation of the response. Figure 12 shows the possible outcomes of a T
cell receiving one or both of the signals necessary for activation. Engagement
of the TCR with Ag/MHC but no co-simulation results in anergy. Engagement of
only the co-stimulatory molecule has no effect. Engagement of TCR with Ag/MHC
and co-stimulatory molecules with their ligand results in activation. Engagement
of the TCR with Ag/MHC and engagement of B7 ligand with CTLA-4, molecules
similar to CD28, results in down-regulation of the response. CTLA-4/B7
interaction sends an inhibitory signal to the T cell rather than an activating
signal. This is one of the ways that immune responses are regulated. CTLA-4 is
expressed on T cells later in an immune response and this helps to turn off the
response.
Key Steps in T cell Activation
-
APC must process and present peptides to T cells
-
T cells must receive a co-stimulatory signal - usually from CD28/B7
-
Accessory adhesion molecules must help to stabilize the binding of T cells
and APC. (CD4/class II MHC, CD8/classs I MHC, LFA-1/ICAM-1 and CD2/LFA-3)
-
Signals from cell surface must be transmitted to the nucleus via second
messengers
-
Cytokines, including IL-2, must help drive cell division
|
TABLE 4
IMPORTANT ACCESSORY MOLECULES |
|
T cell molecule |
Ligand on
second cell |
|
CD4 on helper T cells |
class II
MHC molecules |
|
CD8 on cytotoxic T cells |
class
I MHC molecules |
|
LFA-2 (CD2) |
LFA-3 |
|
LFA-1 |
ICAM-1, ICAM-2 |
|
LFA = Leukocyte
Function-associated Antigen |
|
ICAM = Intercellular Adhesion
Molecule |
|
|

 |
 Return to the Immunology Section of Microbiology and Immunology On-line
Return to the Immunology Section of Microbiology and Immunology On-line
This page last changed on
Thursday, September 14, 2017
Page maintained by
Richard Hunt
|

 Figure 1
Figure 1 Figure 3 Most variability in amino acids at different positions along the alpha
chain of class I MHC molecules occurs in the alpha 1 and alpha 2
regions. The greatest polymorphism is found for amino acids that line
the wall and floor of the groove that binds the peptides
Figure 3 Most variability in amino acids at different positions along the alpha
chain of class I MHC molecules occurs in the alpha 1 and alpha 2
regions. The greatest polymorphism is found for amino acids that line
the wall and floor of the groove that binds the peptides Figure 4
Figure 4 Figure 6 MHC class II molecules comprise two non-identical peptides (alpha and
beta) which are non-covalently associated and traverse the plasma
membrane with the N terminus to the outside of the cell. The
domains closest to the membrane in each chain are structurally
related to immunoglobulins. With the exception of the alpha 1
domain, all domains are stabilized by disulfide bridges (red).
Both the alpha and beta chains are glycosylated. The beta chain is
shorter than the alpha chain (beta mol. wt = 28,000) and contains
the alloantigenic sites. There is some polymorphism in the alpha
chain of some MHC II molecules
Figure 6 MHC class II molecules comprise two non-identical peptides (alpha and
beta) which are non-covalently associated and traverse the plasma
membrane with the N terminus to the outside of the cell. The
domains closest to the membrane in each chain are structurally
related to immunoglobulins. With the exception of the alpha 1
domain, all domains are stabilized by disulfide bridges (red).
Both the alpha and beta chains are glycosylated. The beta chain is
shorter than the alpha chain (beta mol. wt = 28,000) and contains
the alloantigenic sites. There is some polymorphism in the alpha
chain of some MHC II molecules Figure 7
Figure 7 Figure 8
Figure 8 Structure of a crystal structure of a complex of a human T cell
receptor, influenza Ha Antigen Peptide and an MHC Class II Molecule.
The alpha and beta chains of the MHC II molecules are in dark and
light blue. The T cell receptor is in yellow and green. The influenza
peptide is in gray. Hennecke, J., Carfi, A., Wiley, D.
C. MMDB Id: 14648 PDB Id: 1FYT. Image prepared using RasMol
Structure of a crystal structure of a complex of a human T cell
receptor, influenza Ha Antigen Peptide and an MHC Class II Molecule.
The alpha and beta chains of the MHC II molecules are in dark and
light blue. The T cell receptor is in yellow and green. The influenza
peptide is in gray. Hennecke, J., Carfi, A., Wiley, D.
C. MMDB Id: 14648 PDB Id: 1FYT. Image prepared using RasMol Figure 10
Figure 10 Figure 11
Figure 11 









